Microbiome confounders and quantitative profiling challenge predicted microbial targets in colorectal cancer development
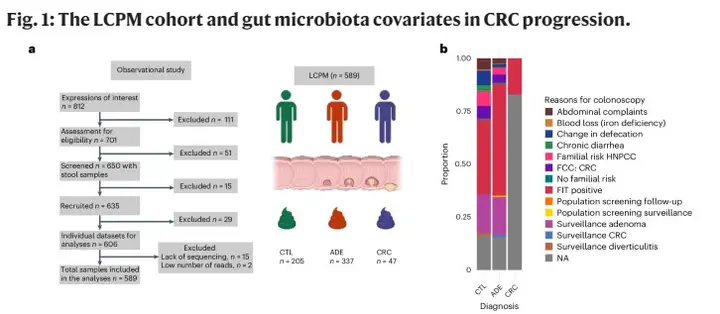 Image credit: Nature Medicine
Image credit: Nature Medicine
Abstract
Despite substantial progress in cancer microbiome research, recognized confounders and advances in absolute microbiome quantification remain underused; this raises concerns regarding potential spurious associations. Here we study the fecal microbiota of 589 patients at different colorectal cancer (CRC) stages and compare observations with up to 15 published studies (4,439 patients and controls total). Using quantitative microbiome profiling based on 16S ribosomal RNA amplicon sequencing, combined with rigorous confounder control, we identified transit time, fecal calprotectin (intestinal inflammation) and body mass index as primary microbial covariates, superseding variance explained by CRC diagnostic groups. Well-established microbiome CRC targets, such as Fusobacterium nucleatum, did not significantly associate with CRC diagnostic groups (healthy, adenoma and carcinoma) when controlling for these covariates. In contrast, the associations of Anaerococcus vaginalis, Dialister pneumosintes, Parvimonas micra, Peptostreptococcus anaerobius, Porphyromonas asaccharolytica and Prevotella intermedia remained robust, highlighting their future target potential. Finally, control individuals (age 22–80 years, mean 57.7 years, standard deviation 11.3) meeting criteria for colonoscopy (for example, through a positive fecal immunochemical test) but without colonic lesions are enriched for the dysbiotic Bacteroides2 enterotype, emphasizing uncertainties in defining healthy controls in cancer microbiome research. Together, these results indicate the importance of quantitative microbiome profiling and covariate control for biomarker identification in CRC microbiome studies.